pFSA is a phagemid vector designed for high-level expression and display of antibody fragment-pIII fusion protein. The Display level depends on phagemid vector as well as helper phage. pFSA significantly increased the expression level of scFv-pIII expression level (Figure 1) and display level (Figure 2).
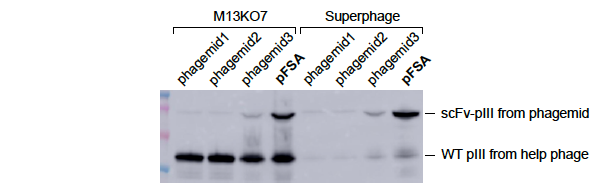
(samples: bacterial lysates)
Figure 1. pFSA significantly increased the expression level of scFv-pIII fusion protein. TG1 cells were transformed with 4 different phagemid vectors phagemid1, phagemid2, phgemid3, and pFSA respectively. All of them expressed the same scFv-pIII fusion protein. The cells were cultured in 2YT media containing 2% glucose and 100ug/ml carbenicillin until OD600 ~0.5. The culture was divided equally and infected by M13KO7 and Superphage respectively. Shake at 60rpm for 15min and then at 250rpm for 45min. Change media to 2YT containing 500ug/ml IPTG and 100ug/ml carbenicillin and 50ug/ml kanamycin. Shake at 250rpm for 5 hours. The bacterial lysates were analyzed by western blot with anti-pIII mAb to detect the expression level of wild-type and fusion pIII. The result showed that (1) pFSA significantly increased the expression of scFv-pIII fusion protein compared to phagemid1, phagemid2, and phgemid3. (2) Superphage dramatically decreased the expression of wild-type pIII expression compared to M13KO7.
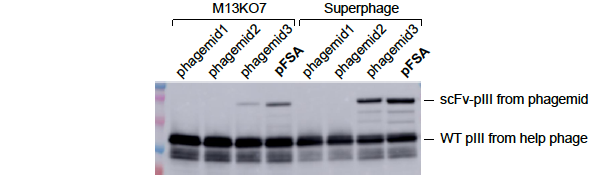
(samples: phage lysates)
Figure 2. pFSA significantly increased the display level of scFv-pIII fusion protein on the surface of M13 phage. TG1 cells were transformed with 4 different phagemid vectors phagemid1, phagemid2, phgemid3, and pFSA respectively. All of them expressed the same scFv-pIII fusion protein. The cells were cultured in 2YT media containing 2% glucose and 100ug/ml carbenicillin until OD600 ~0.5. The culture was divided equally and infected by M13KO7 and Superphage respectively. Shake at 60rpm for 15min and then at 250rpm for 45min. Change media to 2YT containing 500ug/ml IPTG and 100ug/ml carbenicillin and 50ug/ml kanamycin. Shake overnight at 250rpm and 30℃. Phages were purified from bacterial supernatant, and analyzed by western blot with anti-pIII mAb to determine scFv display level. The results showed that (1) the display level of scFv-pIII was the highest with pFSA, and the display level with phagmide1 and phagemid2 were too low to be detected by western blot, and (2) Superphage dramatically increased the display level of scFv-pIII.
In phagemid/helper phage system, each E. coli contains two kinds of pIII for phage assembly (e.g. wild-type pIII derived from the helper phage and antibody-pIII fusion protein derived from the phagemid), and two kinds of genomes for packaging (e.g. ampicillin-resistant ssDNA derived from phagemid and kanamycin-resistant ssDNA derived from helper phage). Therefore, there are four kinds of virus particles in an antibody phage library: (1) phagemid genome and antibody-pIII fusion protein (the desired result). (2) phagemid genome and wild type pIII (unable to bind the target and will be washed away during the panning). (3) phage genome and antibody-pIII fusion protein (possible to bind a target but cannot be amplified). (4) phage genome and wild type pIII (the helper phage itself). In theory, the mutated packaging signal in helper phage should significantly reduce the number of helper phage particles during the library assembly. However, the number of phage particles can sometimes equal, or exceed, the number of phagemid particles, which can significantly compromise subsequent selections[1].
To analyze the ratio of phagemid particles to phage particles, bacteria were infected with the virus particles. Spread the infected bacteria on carbenicillin plates to calculate the titer of phagemid particles (Table 1), and on kanamycin plates to calculate the titer of phage particles (Table 2). The ratio of phagemid particles to phage particles depends on both helper phage and phagemid vector. In the pFSA/Superphage system, the ratio of phagemid particles to phage particles was 500000:1 (Table 3), and almost all the virus particles are phagemid particles.
Table1. Titer of phagemid particles (cfu/ml)
|
|
phagemid1 |
phagemid2 |
phagemid3 |
pFSA |
|
M13KO7 |
2.0E+11 |
5.0E+10 |
1.5E+11 |
1.0E+11 |
|
Superphage |
1.0E+11 |
2.0E+11 |
1.0E+11 |
1.0E+11 |
Table2. Titer of phage particles (cfu/ml)
|
|
phagemid1 |
phagemid2 |
phagemid3 |
pFSA |
|
M13KO7 |
2.0E+11 |
1.0E+11 |
2.5E+10 |
1.5E+10 |
|
Superphage |
3.0E+11 |
7.5E+10 |
4.0E+7 |
2.0E+5 |
Table3. The ratio of phagemid particles to phage particles
|
|
phagemid1 |
phagemid2 |
phagemid3 |
pFSA |
|
M13KO7 |
1:1 |
1:2 |
6:1 |
7:1 |
|
Superphage |
1:3 |
1:3.75 |
250:1 |
500000:1 |
Table4. Specifications of pFSA
Vector category | phagemid |
Replication origin | f1, ColE1 |
Promoter | lac |
Signal peptide | pelB |
Fusion protein | Full length pIII |
Selection | ampicillin |
Quantity | 20ug, Lyophilized |
Reconstitution | Spin down, add 40ul of nuclease-free water/TE/EB buffer, shaking for 30min at 50°C until completely dissolved. Spin down again. |
Amplification | Transformation into NEB stable competent cells (#C3040), maxiprep. |
Short-term Storage | In nuclease-free water or TE, at 4°C |
Long-term Storage | Lyophilized DNA at -20°C; E. coli glycerol stock, -80°C |
References
1. Chasteen L, Ayriss J, Pavlik P, et al. Eliminating helper phage from phage display. Nucleic Acids Res. 2006; 34(21): e145.
Map of pFSA
forword.png)
Sequences are available upon order and request.
Generation of synthetic antibody phage libraries
Library synthesis
1. CDR cassettes were synthesized as single-stranded DNA using trinucleotide “trimers” at the randomized positions.
2. Conversion of ssDNA to dsDNA using reverse primers
ssDNA (100uM) | 10 x 1ul |
Reverse primers (100uM) | 10 x 2ul |
2 x PCR mix (NEB #M0494) | 10 x 50ul |
H2O | 10 x 47ul |
Total | 10 x 100ul |
98℃, 1min; 60℃, 30min | |
3. DNA purification by DNA clean and concentrator kit (Zymo research #D4030)
Restriction enzyme digestion and purification
1. Digestion
Inserts | Phagemid | |
DNA | 40ug | 40ug |
10 x NEBuffer | 40ul | 40ul |
Restriction enzymes | 400U | 400U |
H2O | To 400ul | To 400ul |
37℃, overnight. | ||
2. Purification DNA by clean and concentrator kit (Zymo research #D4030)
Ligation and purification
1. Ligation
Phagemid | 20ug |
Inserts | Molar ratio of phagemid to insert 1: 6 |
10 x T4 DNA ligase buffer | 50ul |
T4 ligase (NEB #M0202M) | 20ul (40000U) |
H2O | To 500ul |
Room temperature, overnight. | |
2. Purification
ligation | 500ul |
3M Sodium acetate | 50ul |
Glycogen (20mg/ml) | 10ul |
100% isopropanol | 1000ul |
Mix, -20℃, > 1hour | |
Centrifuge at 14000rpm and 4℃ for 20min | |
Decant liquid, add 1ml of 70% ethanol to the pellet, rotate for 10 min at 4℃. | |
Centrifuge at 14000rpm and 4℃ for 10min. Decant the liquid. | |
Again, decant liquid, add 1ml of 70% ethanol to the pellet, rotate for 10 min at 4℃. | |
Centrifuge at 14000rpm and 4℃ for 10min. Decant the liquid. | |
Spin briefly to remove the trace amount of ethanol. | |
Air dry and add 50ul of nuclease-free water to dissolve the DNA pellet. | |
Transformation and production of phage library
1. 50ul of DNA, 350ul of electro-competent TG1 cells, 0.2cm Bio-Rad electroporation cuvette.
2. 2.5kV, 200Ω.
3. Add 20ml of SOC, 37℃, 225rpm, 1hour.
4. Determine the library size: serial dilution and spread on 100ug/ml carbenicillin plates. Incubate overnight at 37℃. The total colonies should be between 1E+9 and 1E+10.
5. Add 2 x YT containing 50ug/ml to the 20ml of culture, OD600 0.3~0.5.
6. 37℃, 225rpm, 1hour.
6. Add Superphage at a multiplicity of infection of 20:1 (phage-to-cell-ratio).
7. Shake for 15min at 60rpm, and then for 45min at 225rpm.
8. Spin down at 5000xg for 10min, decant the supernatant, add 500ml of 2 x YT containing 100ug/ml of carbenicillin and 50ug/ml of kanamycin and 500uM of IPTG.
9. Shake at 250rpm and 30℃ for 24 hours.
Phage library purification
1. 10000xg, 10min at 4°C degree, collect the supernatant and trash the pellet.
2. Add 20g of PEG-8000 (4% w/v) and 15g of NaCl (3% w/v). After the solid is completely dissolved, keep in ice for 60min or overnight at 4°C.
3. Spin at 15000g for 20min at 4C. Discard the supernatant; spin down again and remove the remaining liquid.
4. Resuspend the phage pellet in 20ml of 1x PBS containing 1% BSA and 0.02% NaN3.
5. 15000xg for 10min at 4°C, keep the liquid and trash the pellet.
Storage
1. Short term: 4°C for 1 week.
2. Long term: Adjust the phage concentration to 1E+12 cfu/ml in 1X PBS with 10% glycerol. Aliquot and store at -80℃. Avoid repeated freezing and thawing.
1. This product is available to nonprofit organizations or for-profit companies.
2. Buyers are NOT allowed to transfer or resell this product in any form.
pFSA
- Product Code: PM001
-
$1,098.00
Related Products
M13PK
M13PK is the double-stranded, circular DNA derived from bacteriophage M13 phage, and has the fo..
$598.00
Superphage
Superphage is a derivative of M13KO7. In bacteria, the wildtype pIII level from M13KO7 is much highe..
$2,098.00
Phage Display Package
The phage display package contains 4 items. 1) pFSA is a phagemid vector designed for high-leve..
$3,098.00




